After reviewing some results from uclust and uclust_ref OTU picking, we’ve decided to change the default parameters associated with these OTU pickers in the QIIME pick_otus.py scripts. These changes have the effect of slowing down the OTU pickers, but give better results.
We were noticing in certain cases (notably the gg_otus_6oct2010) that we had representative sequences that were greater than 97% similar. This led to generation of multiple OTUs that should have been a single OTU, or ‘split OTUs’. The changes we’ve made to address this are: increase max_accepts from 8 to 20; increase max_rejects from 32 to 500; increase stepwords from 8 to 20; and increase word_length from 8 to 12. This appears to get around the ‘split OTU’ problem.
Performance comparisons based on 50,000 to 250,000 sequences (median length: 235) are below (Figures 1 and 2). The increase in run time is not much different for uclust (de novo), but is quite a bit higher for uclust_ref because there is more extensive testing against a very large collection of seeds. These comparisons were performed using an Amazon Web Services m2.4xlarge instance with the QIIME EC2 image (modified to use the SVN version of QIIME).
We’ve also changed the default representative sequence picking method from most_abundant to first. This ensures that the uclust seed is taken as the representative sequence for each OTU. Sequence abundance is taken into account upstream in pick_otus.py now (as it has been since QIIME 1.1.0) using the presort by abundance feature which ensures that abundant sequences are more likely to be used as OTU seeds than less abundant sequences. In practice, the difference in what sequence is chosen as the representative is minimal as the most abundant sequence is always the first in each cluster, but can be thrown off if there is a tie for the most abundant sequence within an OTU.
In this process we’ve also re-picked our greengenes OTUs. The new version, gg_otus_29nov2010, is available from the Greengenes site here.
If you’re using QIIME 1.2.0, you can call pick_otus.py –max_accepts 20 –max_rejects 500 –uclust_stable_sort; pick_rep_set.py -m first to get close to the new defaults in QIIME 1.2.0-dev. The –stepwords and –word_length options are currently available only in the svn version of QIIME, and will go into QIIME 1.3.0. If you’re using SVN QIIME and you’d prefer uclust to run faster, you can also set these values back to the 1.2.0 defaults (which are listed below) on the command line or in a parameters file.
Finally, thanks to Les Dethlefsen at Stanford for initial investigations on what these parameter settings should be for OTU picking!
Greg
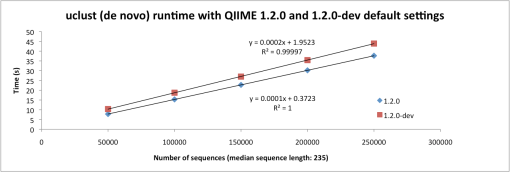
Figure 1: Run time of pick_otus.py -m uclust with QIIME 1.2.0 (blue) and 1.2.0-dev (red) parameters.
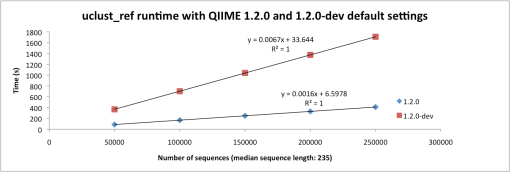
Figure 2: Run time of pick_otus.py -m uclust -C with QIIME 1.2.0 (blue) and 1.2.0-dev (red) parameters.
In summary:
QIIME 1.2.0-dev defaults:
pick_otus:max_accepts 20
pick_otus:max_rejects 500
pick_otus:stepwords 20
pick_otus:word_length 12
pick_rep_set:rep_set_picking_method first
QIIME 1.2.0 defaults:
pick_otus:max_accepts 8
pick_otus:max_rejects 32
pick_otus:stepwords 8
pick_otus:word_length 8
pick_rep_set:rep_set_picking_method most_abundant
